
Nuclear magnetic resonance spectroscopy is a powerful and unique tool for the study of matter in its various states, including solids, liquids, liquid crystals, and biological materials. With the use of experimental and theoretical NMR methods, one can obtain knowledge of the equilibrium structural properties and molecular dynamics of liquid crystals and phospholipid membranes. The latter systems, in particular, exhibit complex and interesting phase diagrams, with a rich variety of nanostructures, including planar lamellar phases, as well as non-lamellar phases involving hexagonally packed nanotubes and phases with cubic symmetry. Various one- and two-dimensional NMR experiments are carried out, in which the spectral lineshapes yield knowledge of the microstructure, whereas the NMR relaxation times afford new insights into the molecular dynamics. A major emphasis involves developing solid-state NMR techniques and relaxation theory for the study of molecular solids and liquid crystals. In this work deuterium (2H) quadrupolar echo spectroscopy is used to characterize the structural properties, relaxation behavior, and molecular dynamics. We are presently utilizing 2H NMR to investigate influences of the bilayer thickness, polyunsaturation, polar head groups, and the incorporation of detergents, cholesterol, and proteins such as rhodopsin on the elastic properties of phospholipid membranes.
Biophysics and biochemistry of membranes
Our studies of lipid-protein interactions have significantly advanced the field of biomembranes. Notably, we have extended the standard model (the fluid mosaic model) found in most biochemistry texts. We discovered how membrane lipids govern the conformational energetics of proteins by our innovation of a flexible surface model (FSM). In our picture, frustration (elastic deformation) of the membrane is counterbalanced by the hydrophobic mismatch of the lipids to the protein surface. Our bold new ideas explain how the tightly regulated lipids affect biological functions of proteins in signal transduction, ion conductance, and metabolite transport. Conformational changes involve elastic remodeling of the lipid bilayer that affects lipid-protein and lipid-peptide interactions (described by differential geometry). Allosteric regulation of membrane proteins is linked to influences of bilayer thickness, nonlamellar-forming lipids, detergents, and osmotic stress that are explained by the FSM as a new paradigm. Currently, my students are investigating how properties such as curvature stress and membrane surface potential govern light activation of rhodopsin. The role of the membrane lipid composition in controlling activation of the G-protein (transducin) is also under study. Investigations of the roles of omega-3 polyunsaturated lipids in visual function and dysfunction are carried out. Our work illuminates how membrane properties underlie key cellular processes involved with signaling, ion channels, and drug transport.
Structure and Dynamics of Proteins Studied by Multidimensional NMR Spectroscopy
Additional work involves extending and applying the above approaches to proteins, to obtain information on their molecular dynamics over a wide range of time scales. As a rule, the relaxation times of proteins depend on the mean-squared amplitudes of the motions of the various functional groups and the associated correlation times. Hence one can deduce information regarding the average structure as well as dynamical properties of the polypeptide backbone and amino acid side chains. Using multidimensional NMR spectroscopy in conjunction with modern NMR relaxation methods, one can thus understand how protein structural and dynamical properties are related to chemical reactivity, and to biological function.
Solid-State NMR Spectroscopy of Membrane Proteins and Nucleic Acids
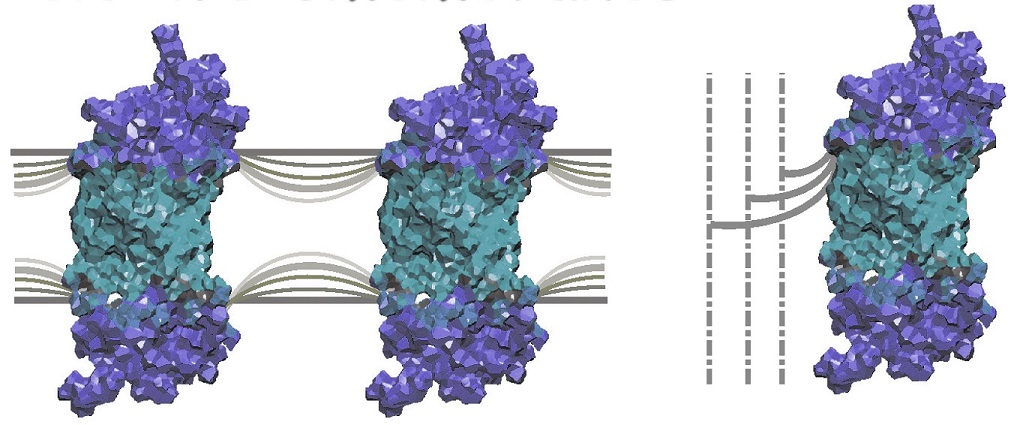
Another major area of research involves the use of solid-state NMR techniques to investigate membrane-associated peptides and integral membrane proteins which are receptors for light, hormones, and neurotransmitters. Our strategy involves deuterium labeling of the ligands for integral membrane proteins in conjunction with 2H NMR spectroscopy of membranes aligned on planar substrates. In this manner, one can obtain information at atomic level resolution regarding the structure and dynamics of the membrane-associated peptides and proteins that cannot be prepared for X-ray diffraction structural studies. Experimental NMR spectra as a function of tilt angle are simulated using theoretical lineshape models, to obtain the bond angles of the various 2H-labeled groups with respect to the membrane. The angular constraints are then used for molecular modeling of the orientation and conformation of the protein-bound ligand within the membrane. As an initial application, we have recently shown how solid-state 2H NMR spectroscopy can be used to study the retinal chromophore of bacteriorhodopsin, a membrane protein involved in solar energy conversion. Future work involves an extension to other integral membrane proteins and receptors. In addition, we are engaged in applying solid-state 2H NMR approaches to DNA, which provides a basis for studying the interactions of nucleic acids with drugs, peptides, and proteins.
Molecular Basis of Vision Studied by Fast Photochemical Reaction Techniques
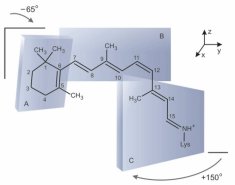
To complement the above NMR studies, we are investigating how equilibrium and dynamical properties of the bilayer lipids influence biological function in the visual system as mediated by the membrane photoreceptor protein rhodopsin. Lipid influences on the photochemistry of rhodopsin in artificial recombinant membranes are studied using rapid kinetic spectrophotometry in real time, together with appropriate kinetic and thermodynamic modeling of the results. Moreover, we are involved with the development and application of surface plasmon resonance (SPR) spectroscopy to study conformational changes of rhodopsin in planar membrane films, and its interactions with other proteins in the biochemical cascade of visual excitation.
Figure made by Karina Martínez Mayorga
Lipid-Protein Interactions and Molecular Modeling of G Protein-Coupled Receptors
Cell membrane receptors are pharmacologically important and constitute about 45% of all molecular drug targets of current therapies. The initial analysis of the human genome has located more than 500 genes involving the rhodopsin-like superfamily of G protein-coupled receptors. In this work, we are utilizing a combination of solid-state NMR spectroscopy and state-of-the-art computer modeling of lipid-protein interactions based on the recently published X-ray structure of rhodopsin. The aim of our research is to remove the constraints due to the protein crystal packing, and model the receptor in a native-like membrane environment. The molecular information from the crystal structure is augmented with data from solid-state 2H NMR spectroscopy, and functional biochemical data. Currently, we are able to identify the structure of the actual hydrophobic lipid/protein interface and locate internal water molecules. By mapping the solvation structure onto the amino acid sequence, we are able to test the performance of hydropathy analysis methods. In the future, the approach can be extended to other G protein-coupled receptors such as the beta-adrenergic receptor as well as the glucagon, oxytocin, and serotonin receptors. Moreover, this work serves as a paradigm for mining structural information from the sequences of membrane proteins embodied in the human genome. It is an important framework for bioinformatic approaches and structural genomics involving membrane proteins.